H/fold.h File Reference
MFE calculations and energy evaluations for single RNA sequences. More...
Go to the source code of this file.
Functions | |
| float | fold_par (const char *sequence, char *structure, paramT *parameters, int is_constrained, int is_circular) |
| Compute minimum free energy and an appropriate secondary structure of an RNA sequence. | |
| float | fold (const char *sequence, char *structure) |
| Compute minimum free energy and an appropriate secondary structure of an RNA sequence. | |
| float | circfold (const char *sequence, char *structure) |
| Compute minimum free energy and an appropriate secondary structure of a circular RNA sequence. | |
| float | energy_of_structure (const char *string, const char *structure, int verbosity_level) |
| Calculate the free energy of an already folded RNA using global model detail settings. | |
| float | energy_of_struct_par (const char *string, const char *structure, paramT *parameters, int verbosity_level) |
| Calculate the free energy of an already folded RNA. | |
| float | energy_of_circ_structure (const char *string, const char *structure, int verbosity_level) |
| Calculate the free energy of an already folded circular RNA. | |
| float | energy_of_circ_struct_par (const char *string, const char *structure, paramT *parameters, int verbosity_level) |
| Calculate the free energy of an already folded circular RNA. | |
| int | energy_of_structure_pt (const char *string, short *ptable, short *s, short *s1, int verbosity_level) |
| Calculate the free energy of an already folded RNA. | |
| int | energy_of_struct_pt_par (const char *string, short *ptable, short *s, short *s1, paramT *parameters, int verbosity_level) |
| Calculate the free energy of an already folded RNA. | |
| void | free_arrays (void) |
| Free arrays for mfe folding. | |
| void | parenthesis_structure (char *structure, bondT *bp, int length) |
| Create a dot-backet/parenthesis structure from backtracking stack. | |
| void | parenthesis_zuker (char *structure, bondT *bp, int length) |
| Create a dot-backet/parenthesis structure from backtracking stack obtained by zuker suboptimal calculation in cofold.c. | |
| void | update_fold_params (void) |
| Recalculate energy parameters. | |
| void | assign_plist_from_db (plist **pl, const char *struc, float pr) |
| Create a plist from a dot-bracket string. | |
| int | LoopEnergy (int n1, int n2, int type, int type_2, int si1, int sj1, int sp1, int sq1) |
| int | HairpinE (int size, int type, int si1, int sj1, const char *string) |
| void | initialize_fold (int length) |
| Allocate arrays for folding . | |
| float | energy_of_struct (const char *string, const char *structure) |
| Calculate the free energy of an already folded RNA. | |
| int | energy_of_struct_pt (const char *string, short *ptable, short *s, short *s1) |
| Calculate the free energy of an already folded RNA. | |
| float | energy_of_circ_struct (const char *string, const char *structure) |
| Calculate the free energy of an already folded circular RNA. | |
Variables | |
| int | logML |
| if nonzero use logarithmic ML energy in energy_of_struct | |
| int | uniq_ML |
| do ML decomposition uniquely (for subopt) | |
| int | cut_point |
| brief set to first pos of second seq for cofolding | |
| int | eos_debug |
| brief verbose info from energy_of_struct | |
Detailed Description
MFE calculations and energy evaluations for single RNA sequences.
This file includes (almost) all function declarations within the RNAlib that are related to MFE folding...
Function Documentation
| float fold_par | ( | const char * | sequence, | |
| char * | structure, | |||
| paramT * | parameters, | |||
| int | is_constrained, | |||
| int | is_circular | |||
| ) |
Compute minimum free energy and an appropriate secondary structure of an RNA sequence.
The first parameter given, the RNA sequence, must be uppercase and should only contain an alphabet 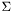 that is understood by the RNAlib
that is understood by the RNAlib
(e.g. 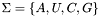 )
)
The second parameter, structure, must always point to an allocated block of memory with a size of at least 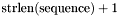
If the third parameter is NULL, global model detail settings are assumed for the folding recursions. Otherwise, the provided parameters are used.
The fourth parameter indicates whether a secondary structure constraint in enhanced dot-bracket notation is passed through the structure parameter or not. If so, the characters " | x < > " are recognized to mark bases that are paired, unpaired, paired upstream, or downstream, respectively. Matching brackets " ( ) " denote base pairs, dots "." are used for unconstrained bases.
To indicate that the RNA sequence is circular and thus has to be post-processed, set the last parameter to non-zero
After a successful call of fold_par(), a backtracked secondary structure (in dot-bracket notation) that exhibits the minimum of free energy will be written to the memory structure is pointing to. The function returns the minimum of free energy for any fold of the sequence given.
- Note:
- OpenMP: Passing NULL to the 'parameters' argument involves access to several global model detail variables and thus is not to be considered threadsafe
- See also:
- fold(), circfold(), model_detailsT, set_energy_model(), get_scaled_parameters()
- Parameters:
-
sequence RNA sequence structure A pointer to the character array where the secondary structure in dot-bracket notation will be written to parameters A data structure containing the prescaled energy contributions and the model details. (NULL may be passed, see OpenMP notes above) is_constrained Switch to indicate that a structure contraint is passed via the structure argument (0==off) is_circular Switch to (de-)activate postprocessing steps in case RNA sequence is circular (0==off)
- Returns:
- the minimum free energy (MFE) in kcal/mol
| float fold | ( | const char * | sequence, | |
| char * | structure | |||
| ) |
Compute minimum free energy and an appropriate secondary structure of an RNA sequence.
This function essentially does the same thing as fold_par(). However, it takes its model details, i.e. temperature, dangles, tetra_loop, noGU, no_closingGU, fold_constrained, noLonelyPairs from the current global settings within the library
Use fold_par() for a completely threadsafe variant
- See also:
- fold_par(), circfold()
- Parameters:
-
sequence RNA sequence structure A pointer to the character array where the secondary structure in dot-bracket notation will be written to
- Returns:
- the minimum free energy (MFE) in kcal/mol
| float circfold | ( | const char * | sequence, | |
| char * | structure | |||
| ) |
Compute minimum free energy and an appropriate secondary structure of a circular RNA sequence.
This function essentially does the same thing as fold_par(). However, it takes its model details, i.e. temperature, dangles, tetra_loop, noGU, no_closingGU, fold_constrained, noLonelyPairs from the current global settings within the library
Use fold_par() for a completely threadsafe variant
- See also:
- fold_par(), circfold()
- Parameters:
-
sequence RNA sequence structure A pointer to the character array where the secondary structure in dot-bracket notation will be written to
- Returns:
- the minimum free energy (MFE) in kcal/mol
| float energy_of_structure | ( | const char * | string, | |
| const char * | structure, | |||
| int | verbosity_level | |||
| ) |
Calculate the free energy of an already folded RNA using global model detail settings.
If verbosity level is set to a value >0, energies of structure elements are printed to stdout
- Note:
- OpenMP: This function relies on several global model settings variables and thus is not to be considered threadsafe. See energy_of_struct_par() for a completely threadsafe implementation.
- Parameters:
-
string RNA sequence structure secondary structure in dot-bracket notation verbosity_level a flag to turn verbose output on/off
- Returns:
- the free energy of the input structure given the input sequence in kcal/mol
| float energy_of_struct_par | ( | const char * | string, | |
| const char * | structure, | |||
| paramT * | parameters, | |||
| int | verbosity_level | |||
| ) |
Calculate the free energy of an already folded RNA.
If verbosity level is set to a value >0, energies of structure elements are printed to stdout
- Parameters:
-
string RNA sequence in uppercase letters structure Secondary structure in dot-bracket notation parameters A data structure containing the prescaled energy contributions and the model details. verbosity_level A flag to turn verbose output on/off
- Returns:
- The free energy of the input structure given the input sequence in kcal/mol
| float energy_of_circ_structure | ( | const char * | string, | |
| const char * | structure, | |||
| int | verbosity_level | |||
| ) |
Calculate the free energy of an already folded circular RNA.
- Note:
- OpenMP: This function relies on several global model settings variables and thus is not to be considered threadsafe. See energy_of_circ_struct_par() for a completely threadsafe implementation.
If verbosity level is set to a value >0, energies of structure elements are printed to stdout
- Parameters:
-
string RNA sequence structure Secondary structure in dot-bracket notation verbosity_level A flag to turn verbose output on/off
- Returns:
- The free energy of the input structure given the input sequence in kcal/mol
| float energy_of_circ_struct_par | ( | const char * | string, | |
| const char * | structure, | |||
| paramT * | parameters, | |||
| int | verbosity_level | |||
| ) |
Calculate the free energy of an already folded circular RNA.
If verbosity level is set to a value >0, energies of structure elements are printed to stdout
- Parameters:
-
string RNA sequence structure Secondary structure in dot-bracket notation parameters A data structure containing the prescaled energy contributions and the model details. verbosity_level A flag to turn verbose output on/off
- Returns:
- The free energy of the input structure given the input sequence in kcal/mol
| int energy_of_structure_pt | ( | const char * | string, | |
| short * | ptable, | |||
| short * | s, | |||
| short * | s1, | |||
| int | verbosity_level | |||
| ) |
Calculate the free energy of an already folded RNA.
If verbosity level is set to a value >0, energies of structure elements are printed to stdout
- Note:
- OpenMP: This function relies on several global model settings variables and thus is not to be considered threadsafe. See energy_of_struct_pt_par() for a completely threadsafe implementation.
- See also:
- make_pair_table(), energy_of_struct_pt_par()
- Parameters:
-
string RNA sequence ptable the pair table of the secondary structure s encoded RNA sequence s1 encoded RNA sequence verbosity_level a flag to turn verbose output on/off
- Returns:
- the free energy of the input structure given the input sequence in 10kcal/mol
| int energy_of_struct_pt_par | ( | const char * | string, | |
| short * | ptable, | |||
| short * | s, | |||
| short * | s1, | |||
| paramT * | parameters, | |||
| int | verbosity_level | |||
| ) |
Calculate the free energy of an already folded RNA.
If verbosity level is set to a value >0, energies of structure elements are printed to stdout
- Parameters:
-
string RNA sequence in uppercase letters ptable The pair table of the secondary structure s Encoded RNA sequence s1 Encoded RNA sequence parameters A data structure containing the prescaled energy contributions and the model details. verbosity_level A flag to turn verbose output on/off
- Returns:
- The free energy of the input structure given the input sequence in 10kcal/mol
| void parenthesis_structure | ( | char * | structure, | |
| bondT * | bp, | |||
| int | length | |||
| ) |
Create a dot-backet/parenthesis structure from backtracking stack.
- Note:
- This function is threadsafe
| void parenthesis_zuker | ( | char * | structure, | |
| bondT * | bp, | |||
| int | length | |||
| ) |
Create a dot-backet/parenthesis structure from backtracking stack obtained by zuker suboptimal calculation in cofold.c.
- Note:
- This function is threadsafe
| void assign_plist_from_db | ( | plist ** | pl, | |
| const char * | struc, | |||
| float | pr | |||
| ) |
Create a plist from a dot-bracket string.
The dot-bracket string is parsed and for each base pair an entry in the plist is created. The probability of each pair in the list is set by a function parameter.
The end of the plist is marked by sequence positions i as well as j equal to 0. This condition should be used to stop looping over its entries
This function is threadsafe
- Parameters:
-
pl A pointer to the plist that is to be created struc The secondary structure in dot-bracket notation pr The probability for each base pair
| int LoopEnergy | ( | int | n1, | |
| int | n2, | |||
| int | type, | |||
| int | type_2, | |||
| int | si1, | |||
| int | sj1, | |||
| int | sp1, | |||
| int | sq1 | |||
| ) |
- Deprecated:
- {This function is deprecated and will be removed soon. Use E_IntLoop() instead!}
| int HairpinE | ( | int | size, | |
| int | type, | |||
| int | si1, | |||
| int | sj1, | |||
| const char * | string | |||
| ) |
- Deprecated:
- {This function is deprecated and will be removed soon. Use E_Hairpin() instead!}
| void initialize_fold | ( | int | length | ) |
Allocate arrays for folding
.
- Deprecated:
- {This function is deprecated and will be removed soon!}
| float energy_of_struct | ( | const char * | string, | |
| const char * | structure | |||
| ) |
Calculate the free energy of an already folded RNA.
- Note:
- This function is not entirely threadsafe! Depending on the state of the global variable eos_debug it prints energy information to stdout or not...
- Deprecated:
- This function is deprecated and should not be used in future programs! Use energy_of_structure() instead!
- Parameters:
-
string RNA sequence structure secondary structure in dot-bracket notation
- Returns:
- the free energy of the input structure given the input sequence in kcal/mol
| int energy_of_struct_pt | ( | const char * | string, | |
| short * | ptable, | |||
| short * | s, | |||
| short * | s1 | |||
| ) |
Calculate the free energy of an already folded RNA.
- Note:
- This function is not entirely threadsafe! Depending on the state of the global variable eos_debug it prints energy information to stdout or not...
- Deprecated:
- This function is deprecated and should not be used in future programs! Use energy_of_structure_pt() instead!
- See also:
- make_pair_table(), energy_of_structure()
- Parameters:
-
string RNA sequence ptable the pair table of the secondary structure s encoded RNA sequence s1 encoded RNA sequence
- Returns:
- the free energy of the input structure given the input sequence in 10kcal/mol
| float energy_of_circ_struct | ( | const char * | string, | |
| const char * | structure | |||
| ) |
Calculate the free energy of an already folded circular RNA.
- Note:
- This function is not entirely threadsafe! Depending on the state of the global variable eos_debug it prints energy information to stdout or not...
- Deprecated:
- This function is deprecated and should not be used in future programs Use energy_of_circ_structure() instead!
- Parameters:
-
string RNA sequence structure secondary structure in dot-bracket notation
- Returns:
- the free energy of the input structure given the input sequence in kcal/mol
 1.6.1
1.6.1