H/plot_layouts.h File Reference
Secondary structure plot layout algorithms. More...
Go to the source code of this file.
Defines | |
| #define | VRNA_PLOT_TYPE_SIMPLE 0 |
| Definition of Plot type simple. | |
| #define | VRNA_PLOT_TYPE_NAVIEW 1 |
| Definition of Plot type Naview. | |
| #define | VRNA_PLOT_TYPE_CIRCULAR 2 |
| Definition of Plot type Circular. | |
Functions | |
| int | simple_xy_coordinates (short *pair_table, float *X, float *Y) |
| Calculate nucleotide coordinates for secondary structure plot the Simple way. | |
| int | simple_circplot_coordinates (short *pair_table, float *x, float *y) |
| Calculate nucleotide coordinates for Circular Plot. | |
Variables | |
| int | rna_plot_type |
| Switch for changing the secondary structure layout algorithm. | |
Detailed Description
Secondary structure plot layout algorithms.
c Ronny Lorenz The ViennaRNA Package
Define Documentation
| #define VRNA_PLOT_TYPE_SIMPLE 0 |
Definition of Plot type simple.
This is the plot type definition for several RNA structure plotting functions telling them to use Simple plotting algorithm
| #define VRNA_PLOT_TYPE_NAVIEW 1 |
Definition of Plot type Naview.
This is the plot type definition for several RNA structure plotting functions telling them to use Naview plotting algorithm
| #define VRNA_PLOT_TYPE_CIRCULAR 2 |
Definition of Plot type Circular.
This is the plot type definition for several RNA structure plotting functions telling them to produce a Circular plot
Function Documentation
| int simple_xy_coordinates | ( | short * | pair_table, | |
| float * | X, | |||
| float * | Y | |||
| ) |
Calculate nucleotide coordinates for secondary structure plot the Simple way.
- See also:
- make_pair_table(), rna_plot_type, simple_circplot_coordinates(), naview_xy_coordinates(), PS_rna_plot_a(), PS_rna_plot, svg_rna_plot()
- Parameters:
-
pair_table The pair table of the secondary structure X a pointer to an array with enough allocated space to hold the x coordinates Y a pointer to an array with enough allocated space to hold the y coordinates
- Returns:
- length of sequence on success, 0 otherwise
| int simple_circplot_coordinates | ( | short * | pair_table, | |
| float * | x, | |||
| float * | y | |||
| ) |
Calculate nucleotide coordinates for Circular Plot.
This function calculates the coordinates of nucleotides mapped in equal distancies onto a unit circle.
- Note:
- In order to draw nice arcs using quadratic bezier curves that connect base pairs one may calculate a second tangential point
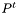 in addition to the actual R2 coordinates. the simplest way to do so may be to compute a radius scaling factor
in addition to the actual R2 coordinates. the simplest way to do so may be to compute a radius scaling factor 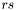 in the interval
in the interval ![$[0,1]$](form_25.png) that weights the proportion of base pair span to the actual length of the sequence. This scaling factor can then be used to calculate the coordinates for
that weights the proportion of base pair span to the actual length of the sequence. This scaling factor can then be used to calculate the coordinates for 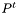 , i.e.
, i.e. ![$ P^{t}_x[i] = X[i] * rs$](form_26.png) and
and ![$P^{t}_y[i] = Y[i] * rs$](form_27.png) .
.
- See also:
- make_pair_table(), rna_plot_type, simple_xy_coordinates(), naview_xy_coordinates(), PS_rna_plot_a(), PS_rna_plot, svg_rna_plot()
- Parameters:
-
pair_table The pair table of the secondary structure x a pointer to an array with enough allocated space to hold the x coordinates y a pointer to an array with enough allocated space to hold the y coordinates
- Returns:
- length of sequence on success, 0 otherwise
Variable Documentation
| int rna_plot_type |
Switch for changing the secondary structure layout algorithm.
Current possibility are 0 for a simple radial drawing or 1 for the modified radial drawing taken from the naview program of Bruccoleri & Heinrich (1988).
- Note:
- To provide thread safety please do not rely on this global variable in future implementations but pass a plot type flag directly to the function that decides which layout algorithm it may use!
 1.6.1
1.6.1