10. Related software¶
Bedtools has been used as an engine behind other genomics software and has been integrated into widely used tools such as Galaxy and IGV. Below is a likely incomplete list. If you know of others, please let us know, or better yet, edit the document on GitHub and send us a pull request. You can do this by clicking on the “Edit and improve this document” link in the lower lefthand corner.
10.1. IGV¶
Bedtools is now integrated into the IGV genome viewer as of IGV version 2.2. We are actively working withe IGV development team to improve and expand this integration. See here and here for details.
10.2. Galaxy¶
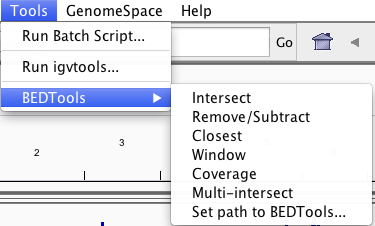
Galaxy has its own tools for working with genomic intervals under the “Operate on Genomic Intervals” section. A subset of complementary Bedtools utilities have also been made available on Galaxy in an effort to provide functionality that isn’t available with the native Galaxy tools.
10.3. Pybedtools¶
Pybedtools is a really fantastic Python library that wraps (and extends upon) the bedtools utilities and exposes them for easy use and new tool development using Python. Pybedtools is actively maintained by Ryan Dale.
10.4. MISO¶
MISO is “a probabilistic framework that quantitates the expression level of alternatively spliced genes from RNA-Seq data, and identifies differentially regulated isoforms or exons across samples.” A subset of the functionality in MISO depends upon bedtools. MISO is developed by Yarden Katz.
10.5. RetroSeq¶
RetroSeq is “a tool for discovery and genotyping of transposable element variants (TEVs) (also known as mobile element insertions) from next-gen sequencing reads aligned to a reference genome in BAM format”. RetroSeq is developed by Thomas Keane. Source code can be obtained on GitHub.
10.6. Intersphinx documentation¶
BEDTools documentation pages are available via Intersphinx (http://sphinx-doc.org/ext/intersphinx.html). To enable this, add the following to conf.py in a Sphinx proejct:
intersphinx_mapping = {‘bedtools’: (‘http://bedtools.readthedocs.org/en/latest/‘, None)}
BEDtools documentation links can then be generated with e.g.:
Table Of Contents
Previous topic
This Page
Quick search
Enter search terms or a module, class or function name.