bedtools: a powerful toolset for genome arithmetic¶
Collectively, the bedtools utilities are a swiss-army knife of tools for a wide-range of genomics analysis tasks. The most widely-used tools enable genome arithmetic: that is, set theory on the genome. For example, bedtools allows one to intersect, merge, count, complement, and shuffle genomic intervals from multiple files in widely-used genomic file formats such as BAM, BED, GFF/GTF, VCF. While each individual tool is designed to do a relatively simple task (e.g., intersect two interval files), quite sophisticated analyses can be conducted by combining multiple bedtools operations on the UNIX command line.
Interesting usage examples¶
To whet your appetite, here are a few examples of ways in which bedtools has been used for genome research. If you have interesteding examples, please send them our way and we will add them to the list.
- Coverage analysis for targeted DNA capture. Thanks to Stephen Turner.
- Measuring similarity of DNase hypersensitivity among many cell types
- Extracting promoter sequences from a genome
- Comparing intersections among many genome interval files
- RNA-seq coverage analysis. Thanks to Erik Minikel.
- Identifying targeted regions that lack coverage. Thanks to Brent Pedersen.
- Calculating GC content for CCDS exons.
- Making a master table of ChromHMM tracks for multiple cell types.
Table of contents¶
Performance¶
As of version 2.18, bedtools is substantially more scalable thanks to improvements we have made in the algorithm used to process datasets that are pre-sorted by chromosome and start position. As you can see in the plots below, the speed and memory consumption scale nicely with sorted data as compared to the poor scaling for unsorted data. The current version of bedtools intersect is as fast as (or slightly faster) than the bedops package’s bedmap which uses a similar algorithm for sorted data. The plots below represent counting the number of intersecting alignments from exome capture BAM files against CCDS exons. The alignments have been converted to BED to facilitate comparisons to bedops. We compare to the bedmap --ec option because similar error checking is enforced by bedtools.
Note: bedtools could not complete when using 100 million alignments and the R-Tree algorithm used for unsorted data owing to a lack of memory.
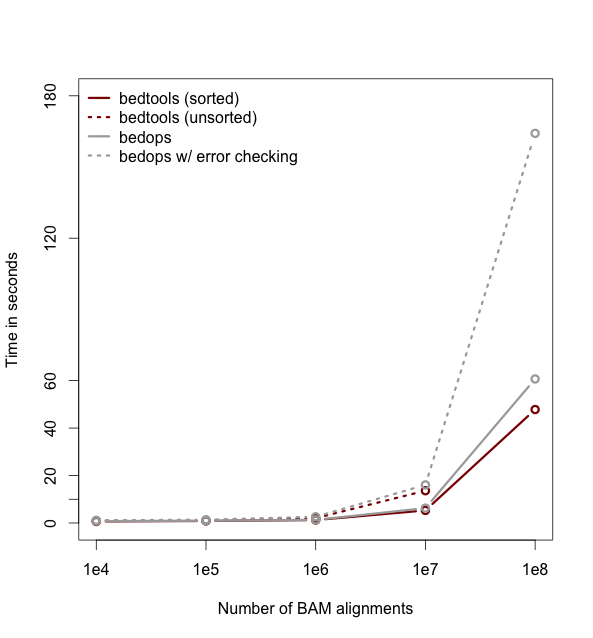
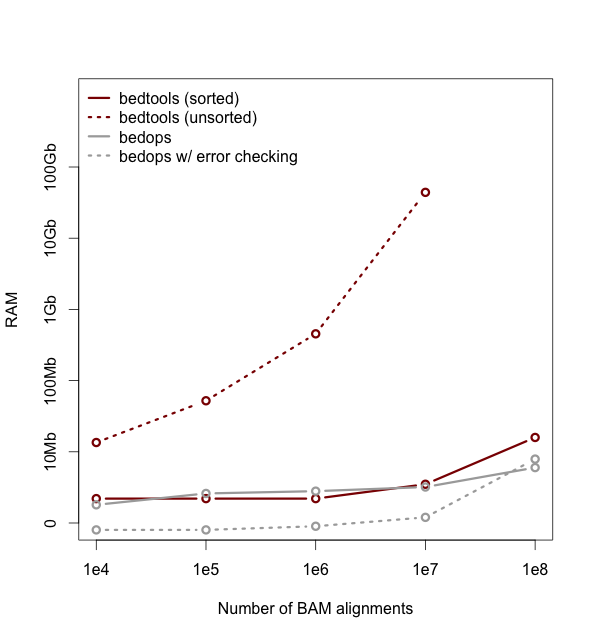
Commands used:
# bedtools sorted
$ bedtools intersect \
-a ccds.exons.bed -b aln.bam.bed \
-c \
-sorted
# bedtools unsorted
$ bedtools intersect \
-a ccds.exons.bed -b aln.bam.bed \
-c
# bedmap (without error checking)
$ bedmap --echo --count --bp-ovr 1 \
ccds.exons.bed aln.bam.bed
# bedmap (no error checking)
$ bedmap --ec --echo --count --bp-ovr 1 \
ccds.exons.bed aln.bam.bed
Brief example¶
Let’s imagine you have a BED file of ChiP-seq peaks from two different experiments. You want to identify peaks that were observed in both experiments (requiring 50% reciprocal overlap) and for those peaks, you want to find to find the closest, non-overlapping gene. Such an analysis could be conducted with two, relatively simple bedtools commands.
# intersect the peaks from both experiments.
# -f 0.50 combined with -r requires 50% reciprocal overlap between the
# peaks from each experiment.
$ bedtools intersect -a exp1.bed -b exp2.bed -f 0.50 -r > both.bed
# find the closest, non-overlapping gene for each interval where
# both experiments had a peak
# -io ignores overlapping intervals and returns only the closest,
# non-overlapping interval (in this case, genes)
$ bedtools closest -a both.bed -b genes.bed -io > both.nearest.genes.txt
License¶
bedtools is freely available under a GNU Public License (Version 2).
Acknowledgments¶
To do.
Mailing list¶
If you have questions, requests, or bugs to report, please email the bedtools mailing list
