ACCCCCUCCU UCCUUGGAUC AAGGGGCUCA A,
using default parameters.
The energy dot plot is an integral part of the folding prediction. Consider the
folding of a short RNA sequence:
ACCCCCUCCU UCCUUGGAUC
AAGGGGCUCA A,
using default parameters.
![]() kcal/mole at
37
kcal/mole at
37![]() ,
so
,
so
![]() rather than 5% of
rather than 5% of ![]() .
A single,
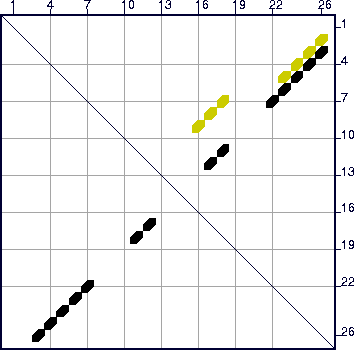
optimal folding is computed. A glance of the energy dot plot , shown in Figure
13, reveals the optimal folding in black dots (symbols), but
another set of yellow dots, indicating base pairs in at least 1 other
suboptimal folding. The default value of `W' (2, from Table
3) is too large for this other folding to be predicted, but
a glance at the dot plot shows that something else is there. When
the sequence is refolded with `W'=0, a second, totally different
folding is predicted. Figure 14 displays these foldings with
individual bases drawn.
.
A single,
optimal folding is computed. A glance of the energy dot plot , shown in Figure
13, reveals the optimal folding in black dots (symbols), but
another set of yellow dots, indicating base pairs in at least 1 other
suboptimal folding. The default value of `W' (2, from Table
3) is too large for this other folding to be predicted, but
a glance at the dot plot shows that something else is there. When
the sequence is refolded with `W'=0, a second, totally different
folding is predicted. Figure 14 displays these foldings with
individual bases drawn.
 |
![\begin{figure}\centering
\subfigure[]{\epsfig{file=1-again_1_zoom.ps,width=0.3\t...
...\subfigure[]{\epsfig{file=1-again_2_zoom.ps,width=0.3\textwidth} }\end{figure}](img123.gif) |
 | Michael Zuker Institute for Biomedical Computing Washington University in St. Louis 1998-12-05 |