![[*]](crossref.png)
The main model to understand is the genewise model (called genewise 21:93
for reasons discussed below). It is this model which the other models
are based on - for the estwise models, by removing the intron generating
part of the models, and for the other genewise algorithms by making
approximations to genewise21:93. A diagramatic representation of genewise21:93
is shown in Figure ![[*]](crossref.png)
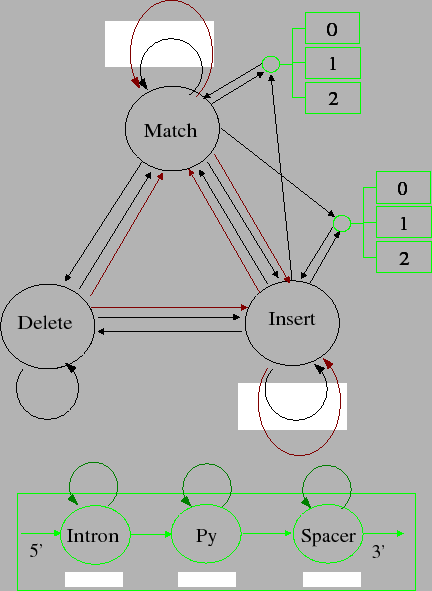 |
The central part of the model is the Match-Insert-Delete trio common to both profile HMMs (such as HMMER models) and the smith waterman model. This trio of states is one model 'position' in the profile HMMs, where each model position contains a Match, Insert and Delete states. This means to interpret the figure of the model in the way the profile HMM models are usually displayed, you have to imagine a series of these states concatonated together. I imagine the model growing as stack of pages out from the figure, each new page being a new position in the profile HMM.
The first addition to the model are the frameshifting transitions, shown in with x4 boxes above them. These occur whenever there is a transition which produces a codon: in effect all transitions that terminate at either match or insert states. There are four frameshifting transitions in each Notice that there are frameshifting transitions from Delete to Match, which is equivalent to saying that a frameshift occurs on the codon just after a run of deletions in the model. It is these sorts of frameshifts that are not well modelled by other algorithms.
The second addition involves the intron emitting states found in the green boxes. Each intron is modelled by having 5 regions, two of which are fixed length. The five regions are
Notice that there is no branch site, because we could not produce a good enough statistical model for it.
This model can be modelled using 3 states, with the fixed length regions being accommodated using transitions which emitted the appropiate length of sequence.
Each of the intron models must be duplicated 3 times to account for the 3 different phases of introns (each phase being a different placement of the intron relative to the codon), so we need to duplicated these 3 states at least 3 times. In addition, if this intron lies in an insert state, ie, the surrounding protein sequence in the exons are being produced by an insert state in the underlying protein profile HMM, so we have to maintain that information across the intron. This means that we need to duplicate the intron states 6 times in total: 3 times for the different phases and twice on top of that for the different protein states this intron could lie in.