5.1.8. cluster¶
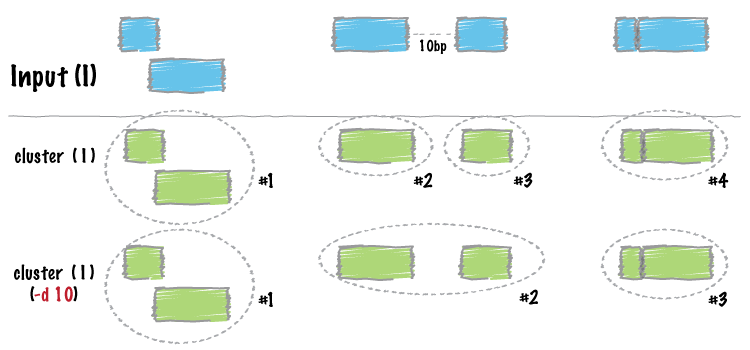
Similar to merge, cluster report each set of overlapping or “book-ended” features in an interval file. In contrast to merge, cluster does not flatten the cluster of intervals into a new meta-interval; instead, it assigns an unique cluster ID to each record in each cluster. This is useful for having fine control over how sets of overlapping intervals in a single interval file are combined.
Note
bedtools cluster requires that you presort your data by chromosome and then by start position (e.g., sort -k1,1 -k2,2n in.bed > in.sorted.bed for BED files).
See also
5.1.8.1. Usage and option summary¶
Usage:
bedtools cluster [OPTIONS] -i <BED/GFF/VCF>
(or):
clusterBed [OPTIONS] -i <BED/GFF/VCF>
| Option | Description |
|---|---|
| -s | Force strandedness. That is, only cluster features that are the same strand. By default, this is disabled. |
| -d | Maximum distance between features allowed for features to be clustered. Default is 0. That is, overlapping and/or book-ended features are clustered. |
5.1.8.2. Default behavior¶
By default, bedtools cluster collects overlapping (by at least 1 bp) and/or bookended intervals into distinct clusters. In the example below, the 4th column is the cluster ID.
$ cat A.bed
chr1 100 200
chr1 180 250
chr1 250 500
chr1 501 1000
$ bedtools cluster -i A.bed
chr1 100 200 1
chr1 180 250 1
chr1 250 500 1
chr1 501 1000 2
5.1.8.3. -s Enforcing “strandedness”¶
The -s option will only cluster intervals that are overlapping/bookended and are on the same strand.
$ cat A.bed
chr1 100 200 a1 1 +
chr1 180 250 a2 2 +
chr1 250 500 a3 3 -
chr1 501 1000 a4 4 +
$ bedtools cluster -i A.bed -s
chr1 100 200 a1 1 + 1
chr1 180 250 a2 2 + 1
chr1 501 1000 a4 4 + 2
chr1 250 500 a3 3 - 3
5.1.8.4. -d Controlling how close two features must be in order to cluster¶
By default, only overlapping or book-ended features are combined into a new feature. However, one can force cluster to combine more distant features with the -d option. For example, were one to set -d to 1000, any features that overlap or are within 1000 base pairs of one another will be clustered.
$ cat A.bed
chr1 100 200
chr1 501 1000
$ bedtools cluster -i A.bed
chr1 100 200 1
chr1 501 1000 2
$ bedtools cluster -i A.bed -d 1000
chr1 100 200 1
chr1 501 1000 1
