5.1.33. slop¶
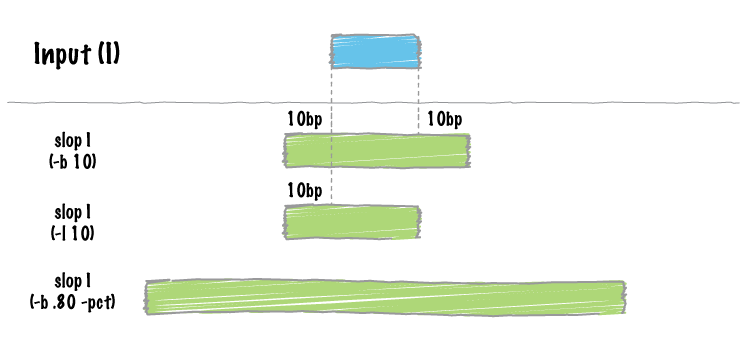
bedtools slop will increase the size of each feature in a feature file by a user-defined number of bases. While something like this could be done with an awk '{OFS="\t" print $1,$2-<slop>,$3+<slop>}', bedtools slop will restrict the resizing to the size of the chromosome (i.e. no start < 0 and no end > chromosome size).
Note
In order to prevent the extension of intervals beyond chromosome boundaries, bedtools slop requires a genome file defining the length of each chromosome or contig.
See also
5.1.33.1. Usage and option summary¶
Usage:
bedtools slop [OPTIONS] -i <BED/GFF/VCF> -g <GENOME> [-b or (-l and -r)]
(or):
slopBed [OPTIONS] -i <BED/GFF/VCF> -g <GENOME> [-b or (-l and -r)]
| Option | Description |
|---|---|
| -b | Increase the BED/GFF/VCF entry by the same number base pairs in each direction. Integer. |
| -l | The number of base pairs to subtract from the start coordinate. Integer. |
| -r | The number of base pairs to add to the end coordinate. Integer. |
| -s | Define -l and -r based on strand. For example. if used, -l 500 for a negative-stranded feature, it will add 500 bp to the end coordinate. |
| -pct | Define -l and -r as a fraction of the feature’s length. E.g. if used on a 1000bp feature, -l 0.50, will add 500 bp “upstream”. Default = false. |
| -header | Print the header from the input file prior to results. |
5.1.33.2. Default behavior¶
By default, bedtools slop will either add a fixed number of bases in each direction (-b) or an asymmetric number of bases in each direction with -l and -r.
$ cat A.bed
chr1 5 100
chr1 800 980
$ cat my.genome
chr1 1000
$ bedtools slop -i A.bed -g my.genome -b 5
chr1 0 105
chr1 795 985
$ bedtools slop -i A.bed -g my.genome -l 2 -r 3
chr1 3 103
chr1 798 983
However, if the requested number of bases exceeds the boundaries of the chromosome, bedtools slop will “clip” the feature accordingly.
$ cat A.bed
chr1 5 100
chr1 800 980
$ cat my.genome
chr1 1000
$ bedtools slop -i A.bed -g my.genome -b 5000
chr1 0 1000
chr1 0 1000
5.1.33.3. -s Resizing features according to strand¶
bedtools slop will optionally increase the size of a feature based on strand.
For example:
$ cat A.bed
chr1 100 200 a1 1 +
chr1 100 200 a2 2 -
$ cat my.genome
chr1 1000
$ bedtools slop -i A.bed -g my.genome -l 50 -r 80 -s
chr1 50 280 a1 1 +
chr1 20 250 a2 2 -
5.1.33.4. -pct Resizing features by a given fraction¶
bedtools slop will optionally increase the size of a feature by a user-specific fraction.
For example:
$ cat A.bed
chr1 100 200 a1 1 +
$ bedtools slop -i A.bed -g my.genome -b 0.5 -pct
chr1 50 250 a1 1 +
$ bedtools slop -i a.bed -l 0.5 -r 0.0 -pct -g my.genome
chr1 50 200 a1 1 +
5.1.33.5. -header Print the header for the A file before reporting results.¶
By default, if your A file has a header, it is ignored when reporting results. This option will instead tell bedtools to first print the header for the A file prior to reporting results.
